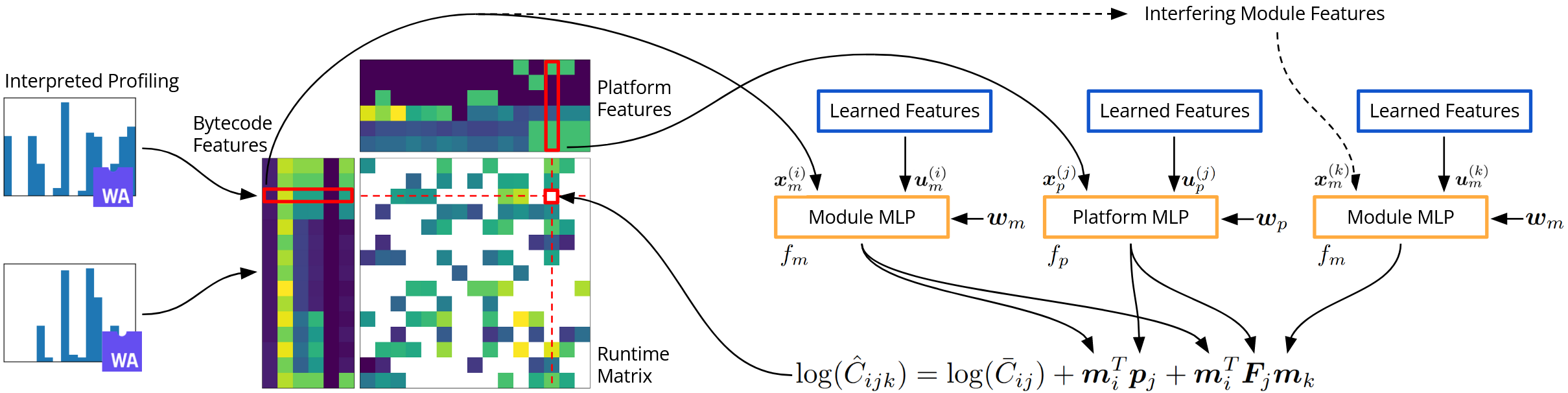
Code and dataset for Interference-Aware Edge Runtime Prediction with Conformal Matrix Completion.
-
Setup: Assuming you have a nvidia GPU, you can simply install all dependencies with conda and poetry:
conda create -n pitot python=3.12 poetry install
- The processed dataset used in the paper is included in the
data/folder.
- The processed dataset used in the paper is included in the
-
Train Models: To replicate the experiments shown in the paper:
make splits make experiments
make experimentscallspython manage.py train, which will automatically run all experiments from the specified list which are not present inresults/. Note that each method result folder will contain aconfig.jsonwith all hyperparameters used.- See
pitot/_presets.pyfor the programmatically generated list of experiments. - This assumes that
pythonpoints to the environment python whichpoetry installwas run inside; if this is not the case, you can also modify the makefile withPYTHON=your/python/bin.
-
Evaluate: The results are evaluated summarized to make them easier to analyze later:
make evaluate -j16 # or some other number of jobs make alias
- This runs on CPU, so can be run simultaneously while training is in progress.
- Some runs are shared between multiple ablations, so are
alias'd together for convenience withmake alias.
-
Analyze: The plots shown in the paper can be generated with
make figures
See here for the benchmarks used to collect our dataset.
Dataset conventions:
t: float[N]: observed runtime (execution time), in seconds.i_{axis}: int[N]: index along each axis into matrix/tensor axes.d_{axis}: float[axis:len, axis:features]: side information for each axis.n_{axis}: str[axis:len]: names corresponding to values in each axis.f_{axis}: str[axis:features]: feature names corresponding to side information for each axis.
Dataset entry names:
data.npz:workload,platform; containsd_{},n_{},f_{}data/metadata.if2.npz:workload,platform,interference0if3.npz:workload,platform,interference0,interference1if4.npz:workload,platform,interference0,interference1,interference2
Python modules:
prediction: core resuable library runtime prediction functionality.pitot: implementation of pitot and other baselines shown in our paper.- Core python code is type-annotated wherever possible, and can be statically typechecked with mypy.
Scripts:
preprocess.py/preprocess: data preprocessing steps to turn raw data into.npzdataset files.manage.py/scripts: main split, training, and evaluation scripts.plot.py/plot: scripts for drawing the figures shown in the paper.
Data files:
data: our dataset is included in this repository directly.splits: this directory is created bymake splits.
Result files:
results: outputs of each experiment are saved here. Has the following structure:results/ method/path/ # can be an arbitrary number of levels config.json # configuration parameters used for training 0.1/ # data split size 0.npz # training log for replicate 0 0.pkl # weights for replicate 0 ... ... ...summary: output of method evaluation.- Each
results/method/path/is turned into onesummary/method/path.npz. - Entries are stacked with data split size as the first axis and the replicate as the second axis (on top of any remaining axes, e.g. target quantile)
- Each